Automated assembly of cell-type-specific mechanisms for the regulation of pain and inflammation with INDRA
 INDRA integrates multiple text-mining systems and pathway databases to
automatically extract mechanistic knowledge from the biomedical literature and
through a process of knowledge assembly, build
executable models and causal networks. Based on profiling and perturbational
data, these models can be contextualized to be cell-type specific and used to
explain experimental observations or to make predictions.
INDRA integrates multiple text-mining systems and pathway databases to
automatically extract mechanistic knowledge from the biomedical literature and
through a process of knowledge assembly, build
executable models and causal networks. Based on profiling and perturbational
data, these models can be contextualized to be cell-type specific and used to
explain experimental observations or to make predictions.
In the context of the DARPA Panacea program,
the INDRA team at the
Laboratory of Systems Pharmacology, Harvard Medical School
is working on understanding the regulation of pain and inflammation with the
goal of finding new therapeutics using INDRA.
- Ion-channel mechanism knowledge base
- Ion-channel inhibitor search
- A self-updating model of pain mechanisms
- Neuro-immune interactome
- General applications
- Funding
Ion-channel mechanism knowledge base
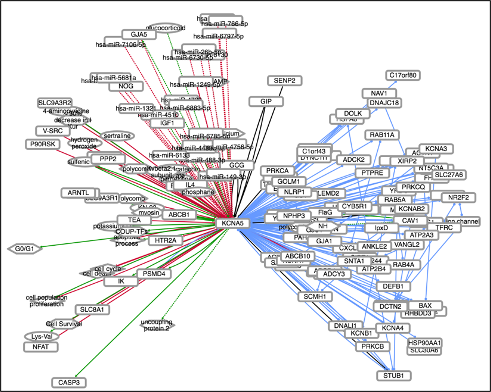
We used INDRA to assemble all mechanisms that 65 ion-channels that are particularly important for nociception are involved in.
Each ion-channel’s interactions can be browsed as networks at
NDEx.
And the literature evidence can be inspected on the pages linked to below. These
pages alsow support expert curation of statements which are fed back to INDRA
to improve the models (see curation tutorial
here.
CACNA1A
CACNA1B
CACNA1C
CACNA1D
CACNA1E
CACNA1F
CACNA1G
CACNA1H
CACNA1I
CACNA1S
HCN1
HCN4
KCNA1
KCNA10
KCNA2
KCNA3
KCNA4
KCNA5
KCNA6
KCNA7
KCNB1
KCNB2
KCNC1
KCNC2
KCNC3
KCNC4
KCND1
KCND2
KCND3
KCNF1
KCNG1
KCNG2
KCNG3
KCNG4
KCNH1
KCNH2
KCNH3
KCNH4
KCNH5
KCNH6
KCNH7
KCNH8
KCNK18
KCNK2
KCNMA1
KCNN1
KCNQ1
KCNQ2
KCNQ3
KCNQ4
KCNQ5
KCNS1
KCNS2
KCNS3
KCNV1
KCNV2
SCN10A
SCN11A
SCN1A
SCN2A
SCN3A
SCN4A
SCN5A
SCN8A
SCN9A
Ion-channel inhibitor search
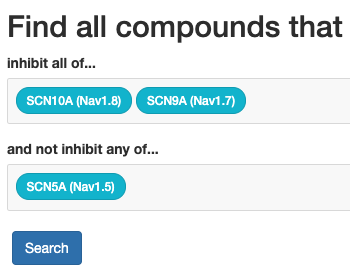 The ion-channel inhibitor search is an application that allows selecting some
ion-channels as “desirable” drug targets and others as “undesirable” drug targets.
The application then searches all INDRA Statements in the ion-channel knowledge
base, combined with the
The ion-channel inhibitor search is an application that allows selecting some
ion-channels as “desirable” drug targets and others as “undesirable” drug targets.
The application then searches all INDRA Statements in the ion-channel knowledge
base, combined with the Small Molecule Suite / Target Affintiy Spectrum
data to find regulators (drugs or other entities that may be relevant) with
the desired properties.
The ion-channel inhibitor search is available here.
A self-updating model of pain mechanisms
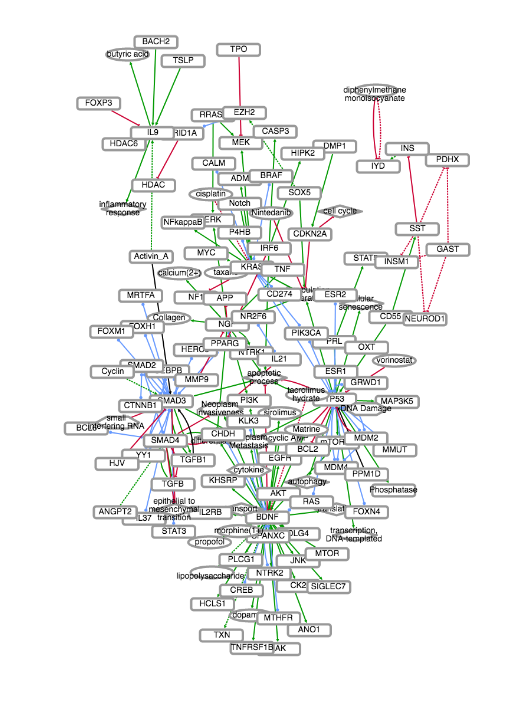 EMMAA (Ecosystem of Machine-maintained Models with Automated Analysis) makes
available a set of
computational models that are kept up-to-date using automated machine reading,
knowledge-assembly, and model generation, integrating new discoveries
immediately as they become available.
EMMAA (Ecosystem of Machine-maintained Models with Automated Analysis) makes
available a set of
computational models that are kept up-to-date using automated machine reading,
knowledge-assembly, and model generation, integrating new discoveries
immediately as they become available.
The EMMAA model representing pain mechanisms can be found
here.
Neuro-immune interactome
Using transcriptional data collected in the Woolf lab
of immune cells and neurons following injury, we are building
cell-type specific models of how immune cells and pain sensing neurons interact.
The latest set of cell-type specific interactions can be browsed on these pages:
DCs
Monocytes
Dermal macrophages
Resident macrophages
M2a macrophages
M2b macrophages
General INDRA applications
There are several applications built on top of INDRA that that are generally applicable to biomedical research and can therefore also be used to study pain mechanisms.
INDRA database: The INDRA database website provides a search interface to find INDRA Statements assembled from the biomedical literature, browse their supporting evidence, and curate any errors. Az example search relevant to Panacea is Object: SCN10A to find entities that regulate the Nav 1.8 ion channel.INDRA network search: The INDRA network search allows finding causal paths, shared regulators, and common targets between two entities. An example search relevant to Panacea is Subject: bradykinin, Object: PKC (seehere).Dialogue.bio: The dialogue.bio website allows launching dedicated human-machine dialogue sessions where you can upload your data (e.g., DE gene lists or gene expression profiles), discuss relevant mechanisms, and build model hypotheses using simple English dialogue. For instance, you could try the following series of questions: “what is Nav1.8?”, “what does it regulate?”, “which of those are transcription factors?”.
Funding
This project is funded under the DARPA Panacea program (HR00111920022).